Dear Steemians,
It has been two weeks since my last Biology roundup article. I had not been able to put out regular round-up posts due to the vacation break I took. It was fantastic to visit France and travel around, I will write up about the amazing culture and food and journey sometime soon.
When I left last time, we saw interesting discoveries/progress made in the field of molecular biology. One article, in particular, caught traction in the scientific community; The article details how an unlikely enterprise between the dengue virus and a gut fungus of the Talaromyces species helps the virus survive in the mosquito gut.
This week there are new and exciting discoveries in the world of molecular biology. I am excited to share with you some articles to kick start the year 2018; I hope I can keep posting the articles along with a review of biological processes weekly.

Weekly round up 7 January 2018
Mangabey's and their resistance to HIV: Novel genomic insights.
When it comes to AIDS, We have been fighting a war against a tricky and sleightful virus. We are into way close to eradication of the virus in humans. A recent study by Guido Silvestri and colleagues may have provided new resources for the fight against aids.
Mangabeys are old world monkeys found in the forests of Senegal and coastal Ghana. It has been known that these monkeys are resilient to infections by Simian Immunodeficiency Virus. Despite high virus load (Viraemia) once infected they are not pathogenic and cause no visible disease. Why is it that a virus so similar to HIV fails to cause disease in these monkeys?
The authors have addressed this using a genome assembly of a captive sooty mangabey. Using genome-wide comparative analyses of AIDS-resistant mangabeys and AIDS-susceptible humans and macaques; They have identified several candidates for host genetic factors influencing susceptibility.
They show that there is a significant divergence in sequences for few key genes from humans and macaques. Genes expressing proteins of Immune system had the major differences. A few of them are;
- C-terminal Frameshift in toll-like receptor-4 gene leads to blunted responses to the TLR4 signalling.
- Structural change in Intercellular adhesion molecule 2(ICAM-2).
- Divergence in sequences of APOBEC3C, BST2. Also in pattern recognition receptors MBL2, CLEC4A/D and CLEC6A and cGAS to name a few. All these are required for a successful HIV infection.
The article by David Plaesch et al., can be found on Nature
Micro-superheroes: How microbiota influences the efficacy of tumour immunotherapy.
Finding two articles concluding the same result is rare but three is something that triggers quite a flurry. In this weeks edition of journal Science; Three papers have shown that microbiota influences the efficacy of anti-PD1 based immunotherapies against cancer.
The article by Gopalakrishnan et al. The authors looked into the oral and intestinal microbiota of patients with metastatic melanoma. The striking observation that they made was, the patients responding to the therapy had an abundance of Faecalibacterium genus.
The article by Routy et al. explored other types of cancers like renal cell carcinoma and non-small cell lung cancer. They outline that the exposure of antibiotics administered during the chemotherapy treatment to ward off any opportunistic infections had a negative correlation to ant-PD-1 treatment response.This suggests that microbial networks may help in the treatment success; In comparison with the faecal microbiota the responders had an abundance of Akkermansia muciniphilia.
Both studies agree that there is a crucial role of certain microbial communities in the responses to the treatment and the outcome success. But they also show that the members of the microbiota vary among cancer types. There are no universal bacterial species that define this response.
The articles can be read on Science in the following links.
Gut microbiome modulates response to anti–PD-1 immunotherapy in melanoma patients
The commensal microbiome is associated with anti–PD-1 efficacy in metastatic melanoma patients
Reprogramming: From Alpha to Beta. How new gene therapy may help against Diabetes.
Diabetes is a global burden. It is estimated that 11-22 million people worldwide are affected by type 1 diabetes(T1D) alone(Source: WHO). T1D usually occurs due to the destruction of pancreatic beta cells; These cells are usually our factories to produce insulin that controls the levels of blood sugar. The treatment of type 1 diabetes usually is to administer artificial/exogenous insulin via injections. Although this has been widely used, There is an exceeding case where the insulin is taken is not useful as there are chances of uptake resistance. Therefore a permanent solution is to reprogram body's own cells.
Researchers at the University of Pittsburgh and the University of Chicago have come up with a novel strategy of reprogramming. Using mouse models and an artificially engineered adeno-associated viruses carrying the capacity to express Pdxq and MafA genes; They were successfully able to reprogram pancreatic alpha cells into beta cells in vivo. The reprogrammed cells delayed diabetes onset and their studies in human alpha cells show that expression of these two genes also reprograms to beta cells.
Graphical Abstract of the paper:
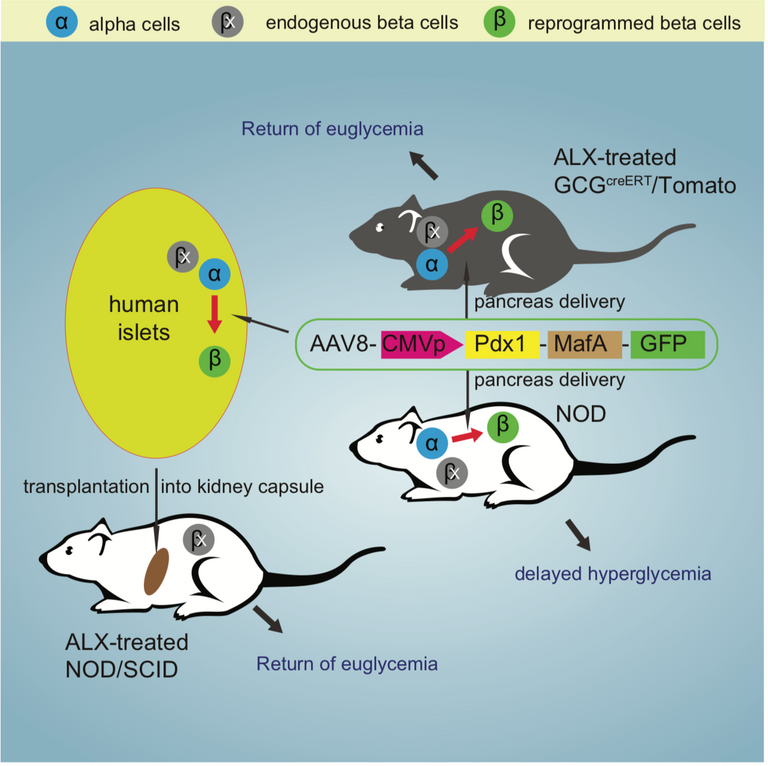
This interesting paper can be accessed on Cell Stem Cell.
Screening Peptides: The hunt for Next-Generation antibiotics continues.
There has been a scarcity of novel and new antibiotics in the market of late. This coupled with an increase in antibiotic resistance has made it more apparent that we need to tackle the problem with a war footing. Antimicrobial peptides have shown promise; There have been demonstrations in the past of their efficacy against gram-negative and gram-positive bacteria. They usually work by disabling the cell wall of bacteria or by other mechanisms not clearly understood.
In a research paper in the Jornal Cell; Researchers Ashely T. Tucker and colleagues describe an Interesting new method to screen several peptides for their bactericidal activity.
The method aptly named SLAY; a short form for Surface Localized Antimicrobial Display has been developed as a platform to allow screening of unlimited numbers of peptides of any length and composition and structure for antimicrobial activity.
Very Briefly;
- A library of random peptides are constructed to be expressed in plasmids and transformed to create an 'input' library.
- These are then transformed into the bacteria to be expressed on the surface; The bacteria expressing the antimicrobial peptides are depleted in this process.
- A next-generation sequencing is performed to identify which of the sequences did not make it in this round in comparison to the 'input'.
- The difference is the sequences that are absent indeed express the peptide. Further screening and verifications are performed to validate the findings.
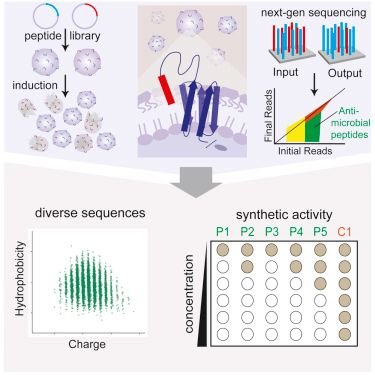
The article can be read at Cell.
Image sources
Sooty Mangabey : San-diego Zoo
Microbiome : Steve Gschmeissner/Science Photo Library.
HeLa Cell: Wiki Commons/NIH.
To Be Continued!
There have been interesting findings on how microbes may help in the treatment of complex diseases and genetic engineering could find its way into daily medical procedures. Science is slow but its effects on humanity are long; It helps us prosper and live. It is rewarding to how scientists are working to solve complex problems.
Participate in the #steemSTEM Community

If you like the post Please consider commenting, upvote and share.

Congratulations @harshameghadri! You received a personal award!
You can view your badges on your Steem Board and compare to others on the Steem Ranking
Vote for @Steemitboard as a witness to get one more award and increased upvotes!