Hello everyone!
A big greeting to all the members of the community #SteemStem. Today I join this great scientific group, in which I hope to collaborate with my works and clinical cases, which I have had to perform in my work as a medical student.
I joined this community a while ago, and not to talk about my career, if not to make music video, which I also like a lot.
Today I will develop a theme that will combine microelectronics and molecular biology, I will talk about DNA matrices.
In recent years, the technological development of bioinformatics equipment has allowed an increase in the number of scientific works focused on complete genomes. Several tools have been developed for its analysis, such as DNA microarray technology, which allows us to study DNA and / or gene expression of thousands of genes in a single experiment.
Let us begin
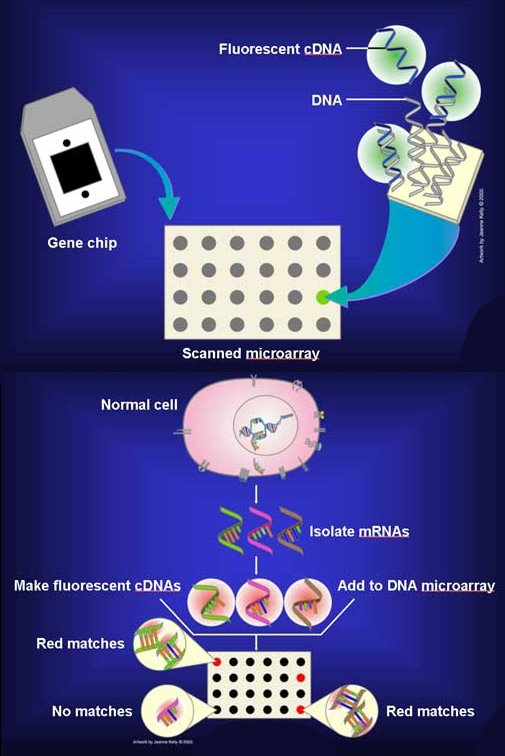
Function of DNA microarrays
This chip is used to identify genes with a differential alteration in different conditions. To detect genes that produce certain diseases by comparing the expression levels between healthy cells and cells that are developing certain types of diseases.
A DNA microarray is a series of DNA probes attached to a solid surface in a regular and preset arrangement. The hybridization of nucleic acids being the basis of the technique.
Its main function is to make an analysis of the level of hybridization that the specific probe and the target molecule have, and they are usually indicated by fluorescence and by means of an image analysis, which indicates the level of expression of the gene.
Types
Spotted Microarrays
This experiment is a little common in obtaining and comparing expression profiles, since two samples are compared: the control RNA and the problematic RNA. The cDNA for each sample is obtained from the RNA, marking each of the samples with a fluorochrome of different color (Cy3 and Cy5). The labeled DNAs will hybridize competitively on the same chip.
DNA oligonucleotides
In the oligonucleotide chips are synthesized on the glass surface of the array, obtaining the synthesis of a number of probes on a single chip (GeneChip). Each probe has a length of about 25 nucleotides, so to complete a gene you need to group several probes, which is called group set. These DNA chips give estimates of the level of expression.
Expression microarray process
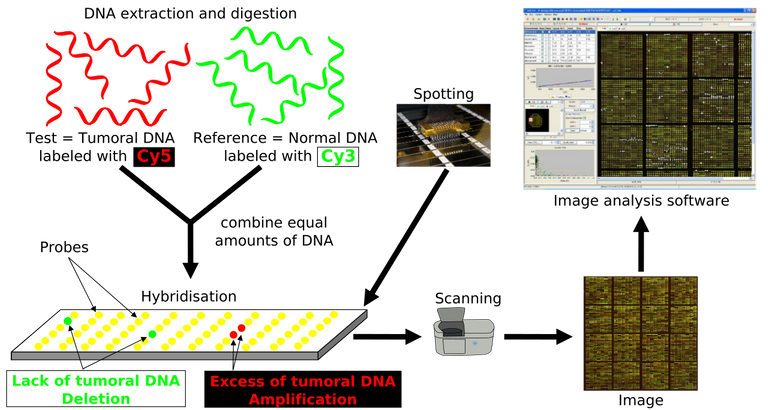
The goal in this experiment is to detect genes that appear in a tissue. This process begins with the extraction of RNA from the sample. The RNA is converted into cDNA by reverse transcriptase and in this process it is labeled with a fluorochrome, that is, with a molecule that will then emit light when it is excited with a suitable laser light.
After the hybridization has occurred, the microarray is read with a laser scanner and 2 images are obtained, one for each fluorochrome used, with points of light whose intensities will vary according to the level of hybridization that has occurred in each objective. These images are processed by software that quantifies the signal of each point (objective) for each fluorochrome (sample) and elaborates a database that will be analyzed with statistical techniques. Often, a superimposed image of the two is made in the form of pseudocolor. One of the images is colored in red and the other in green. In this way, the yellow dots have a high signal in the 2 samples in a similar amount. Red or green dots indicate that the expression of that gene predominates in one of the samples
The RNA is very unstable and degrades in a few minutes, so the tissues must be fresh or frozen immediately after obtaining them.
Application
- Study of genes, which are differentially expressed under diverse conditions (healthy / sick, mutant / wild, treated / untreated).
- Molecular classification in complex diseases. Identification of genes characteristic of a pathology (signature or signature).
- Prediction of response to a treatment.
- Detection of mutations and polymorphisms of a single gene (SNP).
Relevant information
The process of DNA microarrays is not new. Already in the 1980s there were techniques that allowed us to detect genes with a level of expression that were different in biological situations, for example, between tumor cells and normal cells, or between different regions of the brain. Thanks to miniaturization techniques, today many more genes can be visualized at the same time.
With the technique of DNA microarrays we can analyze the entire genome in a single DNA chip.
Conclusion
Although this methodology is still in the embryonic phase with respect to its application in the field of microbiological diagnosis, it presents a series of advantages that make it very attractive and in the future it can be a very valid technique for the diagnosis of diseases infectious
REFERENCES
https://www.cell.com/fulltext/S0092-8674(00)00005-2
https://en.wikipedia.org/wiki/DNA_microarray
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4011503/
Ang, S., Lee, CZ., Peck, K., Sindici, M., Matrubutham, U., Gleeson, MA. y Wang, JT. 2001.
Acid-induced expression in Helicobacter pylori: study in genomic scale by microarray. Infect.
Immun
https://pubs.acs.org/doi/pdf/10.1021/ja991608e
http://www.redalyc.org/html/1812/181221636005/

Thank you for taking the time to read this publication. I hope you have a happy day, see you in the next. Do not forget to leave a comment about what you thought, thank you very much.@vanecsamusic

Hi @vanecsamusic!
Your post was upvoted by utopian.io in cooperation with steemstem - supporting knowledge, innovation and technological advancement on the Steem Blockchain.
Contribute to Open Source with utopian.io
Learn how to contribute on our website and join the new open source economy.
Want to chat? Join the Utopian Community on Discord https://discord.gg/h52nFrV