
The human genome printed out in books that the Sanger method helped to write. If you thought the Lord of the Rings novel series was hard to finish, wait till you get a load of these.
Source: Wikimedia Commons contributors
Intro
This is going to be about another one of those stupendously simple yet Nobel-Prize-winning discoveries that I so love. It's one of the early discoveries that finally allowed scientists to sequence the DNA of the genes they had isolated by cloning (as described here).
Where we left off
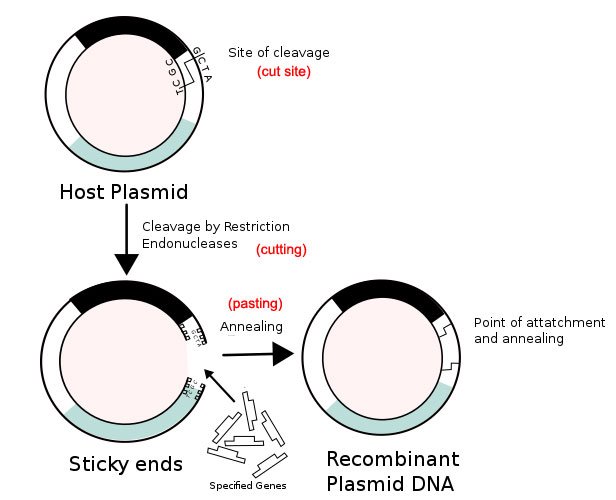
Source: Wikimedia Commons contributors, modified
This is how things stood with plasmids: Scientists had cut the plasmids using special scissors called restriction enzymes; had pasted their gene of interest to them; ligated (joined) the two pieces together; had bacteria take up the plasmids; then the bacteria replicated; and now scientists had lots of genes that they could use for many medical and other purposes, for instance the manufacture of insulin for diabetics.
But they still didn't know how to sequence the letters of those genes. One of the first steps toward doing that was a process called gel electrophoresis. But before doing that, they had to get the genes back out of the plasmids they had sewed them on.
That was a problem. The scientists had made much effort sewing the genes onto the plasmids. How do you reverse that process? It turned out there was a really simple solution to this problem: they used the restriction enzymes again. These enzymes cut the DNA at the very place they had done previously, releasing the now multiplied genes.
And now it was time to measure the size of those genes.
Gel Electrophoresis for length measuring

Size matters!
Also, biology Easter Eggs! Yay!
Source: Wikimedia Commons contributors & PublicDomainPictures, modified
You don't have to be a biologist to know that gel electrophoresis has something to do with gels and electricity. The gel part of the deal is agarose gel. If you are foodie like me, the first question that pops to mind when you hear "gel" is "can I eat it?". You can, and Asia has a long culinary tradition of using agar to make jellies, and vegetarians use it as an alternative to gelatin.[1]

Both are edible, I guess, but one has stuff growing on it that may not agree with you.
Source: Wikimedia Commons contributors a & and b, modified
The DNA fragments migrate through this gel. The reason you need a gel, rather than just water, is to force the fragments to travel slower. Imagine the gel as soil. Smaller worms will encounter less resistance in traveling through the soil, just as a motorcycle can travel faster through traffic. The worm analogy might be amiss because larger worms can use their extra "muscle" to move through the soil faster, whereas DNA all moves at the same speed, because it's not the one providing the energy.
The energy is provided by positively and negatively charged electrodes stuck at opposite ends of the gel floor. DNA is made up of phosphates that have a negative charge, so when the electric current is turned on, the DNA moves toward the positively charged end.

Or click here for a video demonstration.
Source: Wikimedia Commons contributors, modified
The DNA is loaded into wells (holes) in the agarose gel. The first well is reserved for a so-called DNA ladder. That's DNA fragments of known lengths, so that the length of the new DNA can be gauged via comparison.
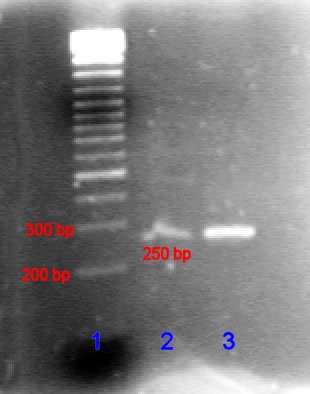
Source: Wikimedia Commons contributors
This is a say-cheese of the agarose gel and tinted DNA (among other things, the DNA can be tinted with fluorescent DNA-binding dye). The line procession at the extreme left is the DNA ladder. The two lines to its right are the products we want to measure, and whose lengths are estimated by comparison to the ladder.
Note again that the smaller DNA fragments will move through the gel more easily, and thus will be closer to the bottom, whereas the lengthier fragments will be closer to the top.
Now you may wonder what all this preamble has to do with sequencing.
Wonder no more!
DNA sequencing: The Sanger way

The protagonist of our story, Frederick Sanger. A late entrée to the story. He was backstage drinking booze. It's made of molten gold Nobel coins.
Source: Wikimedia Commons contributors
Sanger was born to a reasonably wealthy family, proving that it's not just poor kids from Indian villages who excel in the sciences. He was brought up as a Quaker, but he was no quack, as evidenced by his two Nobel Prizes, one of which was for figuring out a way to sequence DNA.
It was a stunningly simple method. It only had one extra ingredient compared to regular DNA replication. Regular DNA replication requires a template DNA, a primer, a DNA polymerase, and the 4 nucleotides G, A, T, and C (I'm abbreviating, they are normally called dGTP, dATP, dTTP, dCTP).
If you add all that to a template DNA, you get a second strand. And here's where the super simple magical ingredient comes in: you add a defective nucleotide. What happens when you add a defective nucleotide, say a defective G? The chain stops. Let's see this more graphically.
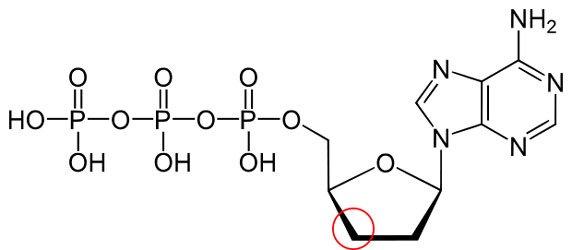
You wouldn't normally know just by looking at it, but this deoxynucleotide (dNTP) is missing an O (oxygen) at the 3' (three prime) position indicated by the red circle, which makes it a dideoxynucleotide (ddNTP). Nice try, pretender!
Source: Wikimedia Commons contributors
What this means is that when the DNA-making process reaches that defective nucleotide, it adds it to the chain, and then stops there, because it doesn't have the O to add new nucleotides to it.
If I've failed to impress you, it's because I haven't made the connection to gel electrophoresation yet.
Let's take a sequence. Say the first several letters of our dear Trp-cage protein that we visited before. Hey, it might be the smallest known protein, but that's no reason not to feature it bigly in this series.
So the first several letters go like this: AACCTGTACATC...
So let's say I add all the necessary ingredients to make new strands. But I also add defective Cs. What will happen? Well, some DNA strands will go A, A, C, then stop, because they added a defective C. That will make a DNA strand of 3bp (three base pairs long). But some strands will take up a non-defective C, and will keep going. They will add another C, which might be defective, or it may not. If it is, they will stop at 4bp. If not, they will continue until the next C.
So we got DNA strands with lengths 3bp, 4bp, 9bp, etc.
Wait, does that mean ... does that mean that the Cs are located at the 3rd, 4th, and 9th places of the DNA sequence?!
It does! Nobel Prize committee you better mint a new golden coin cos this dog ain't barkin'. It's bitin'!
And you can of course do the same with all the other defective nucleotides.
But this article is getting long, so let's do them all. Let's fluorescently label the defective nucleotides with differently-colored dyes, or let's radioactively label them, and see if we can read the sequence.
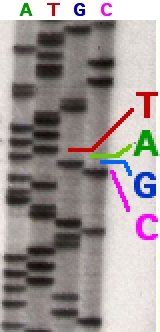
Source: Wikimedia Commons contributors
I spy a line there at the bottom. Does that mean it's shorter than the other bands above? You bet. Does it mean it comes earlier than other bands in the sequence? Yes siree! It's inside the column labeled as T, because of the color — does that mean it's a T? You betcha! Is there a DNA ladder to tell me how many base pairs long it is? Well, not is this pic.
So let's read the sequence. It's TACGAGAT and so on. The strands terminate at each of those sites because they took up a defective nucleotide, giving us all these different lengths. And because we labeled all the nucleotides with different radioactive or fluorescent tags, we know which letter is meant. In fact, because of this, we don't even need 4 columns, we could do it in one.
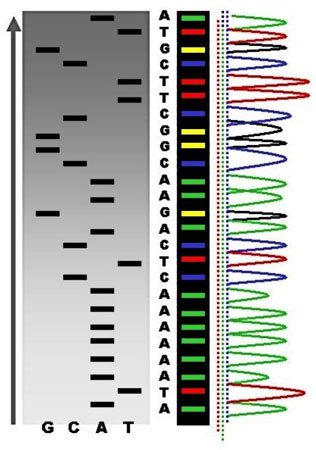
Source: Wikimedia Commons contributors
The colorful column in the middle is where you should be looking!
Outro

Mint me one of these!
Source: Wikimedia Commons contributors
The Sanger method helped sequence the human genome, in a project called The Human Genome Project. See the first pic in this post for a memory-jog. Other, better, newer methods were used primarily, but the Sanger method too was used for smaller sequences.
But what was cutting edge at its time, is now completely outdated. Today you barely even need to move a finger, as the machines do most of the work for you, in what's usually called high-throughput sequencing.[11]
And it's not just ease of use, it's also save of cost. Sequencing a genome used to cost around 100 million $, 15 years back. It now costs little more than 1000 $.[12] Moore's Law ain't got nothing on that! (If you divide 100 million 15 times you're still at the 3000 $ mark.)
But there's one thing at which the Sanger method remains unbeaten to this day: its darn elegant simplicity. Trust me, you don't want me to write a post on high-throughput methods of sequencing!
P.S.
Having demystified the other parts of the term, I don't want to leave you wondering about the phoresis in gel electrophoresis. It means "to carry", or "to bear", from how the electric current carries the electrons, and in our case the dyed DNA along with it, like a surf carries a surfboard.
REFERENCES
1. Wikipedia contributors, "Agar," Wikipedia, The Free Encyclopedia, https://en.wikipedia.org/w/index.php?title=Agar&oldid=820098354 (accessed February 10, 2018).
2. Alberts B, Johnson A, Lewis J, et al. Molecular Biology of the Cell. 4th edition. New York: Garland Science; 2002. Isolating, Cloning, and Sequencing DNA. Available from: https://www.ncbi.nlm.nih.gov/books/NBK26837/
3. Lodish H, Berk A, Zipursky SL, et al. Molecular Cell Biology. 4th edition. New York: W. H. Freeman; 2000. Section 7.3, Identifying, Analyzing, and Sequencing Cloned DNA. Available from: https://www.ncbi.nlm.nih.gov/books/NBK21505/
4. OpenStax, Biology. OpenStax CNX. Mar 22, 2016 http://cnx.org/contents/185cbf87-c72e-48f5-b51e-f14f21b5eabd@10.7. https://cnx.org/contents/GFy_h8cu@10.7:5l844Z38@7/Whole-Genome-Sequencing
5. Wikipedia contributors, "Sanger sequencing," Wikipedia, The Free Encyclopedia, https://en.wikipedia.org/w/index.php?title=Sanger_sequencing&oldid=819406421 (accessed February 10, 2018).
6. Wikipedia contributors, "Dideoxynucleotide," Wikipedia, The Free Encyclopedia, https://en.wikipedia.org/w/index.php?title=Dideoxynucleotide&oldid=629663556 (accessed February 10, 2018).
7. Wikipedia contributors, "Frederick Sanger," Wikipedia, The Free Encyclopedia, https://en.wikipedia.org/w/index.php?title=Frederick_Sanger&oldid=819760419 (accessed February 10, 2018).
8. "The Nobel Prize in Chemistry 1980". Nobelprize.org. Nobel Media AB 2014. Web. 10 Feb 2018. http://www.nobelprize.org/nobel_prizes/chemistry/laureates/1980/
9. Wikipedia contributors, "Gel electrophoresis," Wikipedia, The Free Encyclopedia, https://en.wikipedia.org/w/index.php?title=Gel_electrophoresis&oldid=821103782 (accessed February 10, 2018).
10. Wikipedia contributors, "Electrophoresis," Wikipedia, The Free Encyclopedia, https://en.wikipedia.org/w/index.php?title=Electrophoresis&oldid=814378076 (accessed February 10, 2018).
11. Wikipedia contributors, "DNA sequencing," Wikipedia, The Free Encyclopedia, https://en.wikipedia.org/w/index.php?title=DNA_sequencing&oldid=823930239 (accessed February 10, 2018).
12. The Cost of Sequencing a Human Genome. https://www.genome.gov/sequencingcosts/
Earlier Introduction to Biology episodes:
13: Cutting, Pasting, Cloning, and GMOs
12: How a Boy From an Indian Village Broke the Code of Life and Won the Nobel Prize
11: The Most Beautiful Experiment In Biology
10: The Great GATC-by: The Most Famous Science Paper of the 20th Century
9: The Great Kitchen Blender Experiments: How DNA was proved to be the seat of heredity
8: Finding, Counting, and Ordering Genes Using Incredibly Sophisticated Biomolecular Megatechnology
7: Christmas Disease — Yes, it's real, 100% scientifically proven!
6: The Most Famous All-Nighter in the History of Genetics
5: Mendel's Lucky Number Seven — The law of genetics that almost wasn't
4: How Cells Use Logic To Do The Impossible
3 : Armchair Science — The Discovery of Proteins' Secondary Structure
2 : How Cell Membranes Form Spontaneously
1 : Eduard Buchner: The Man Who Killed Vitalism
steemit.chat or discordsteemSTEM is the go-to place for science on Steemit. Check it out at @steemstem or browse the #steemSTEM tag or chat live at
Being A SteemStem Member
Another great trek down genetics history lane. Sanger sequencing still gets some use in some the core labs I've visited...mostly sequencing PCR fragments and portions of plasmids under 1000 bps. I think it costs under 4 bucks a sample back in Grad school. However, I imagine Sanger wouldn't still be used so frequently if those core labs didn't have so much equipment and history dedicated to them.
both Walter Gilbert and Frederick Sanger for their contributions in determining sequences in DNA. The Maxam–Gilbert sequencing was the first widely adopted method for DNA sequencing, but the Sanger di-deoxy method became the predominant method. The 1980 Nobel prize was also awarded to Paul Berg for his studies on the biochemistry of DNA.
Cheers!@alexander.alexis This is very nice write up on Sanger di-deoxy sequencing. I should point out that the 1980 Nobel Prize in Chemistry was awarded to
Very true. I learned about this while researching for this post, but some leftover phrasing implied otherwise. I now slightly altered the first paragraph to reflect what you said.
Thanks for reading and commenting!
Great article. This is just the beginning and sequencing a genome could become a lot cheaper in future.
It will.
It's amazing what I learn at Steemit daily! Thanks!
@originalworks
Good, nicely explained article :) Easy to follow
Nice article, thanks for sharing such series.